Scientific Overview
DNA RECOMBINATION AND GENOME ENGINEERING IN PLANTS
Our group is analyzing the mechanisms of genomic changes in dicotyledonous plants, which is not only relevant in terms of basic research, but also in terms of biotechnology. We were the first to induce targeted double strand beaks (DSBs) in a plant genome by using rare cutting sequence-specific endonucleases. Using this technique, we could not only elucidate the main mechanisms of DSB repair in plant cells, but we were also the first group worldwide to demonstrate the feasibility of DSB induction for achieving different kinds of genome engineering.
GENOME ENGINEERING USING THE CRISPR/CAS SYSTEM
Today, genome engineering is based on the induction of site-specific DSBs by the use of synthetic nucleases. In our earlier work we used the restriction endonuclease I-SceI with its 18 bp recognition site in transgenic plants. We were able to demonstrate that DSB induction can be used to knock out marker genes by non-homologous end joining (NHEJ) with high efficiency. If two DSBs are set close to each other on the same chromosome, programmed deletions can be obtained. If the two breaks are induced on different chromosomes, reciprocal translocations can be achieved.
We were also able to show that if a DSB is induced during transformation with Agrobacterium tumefaciens, the T-DNA integration occurs more often than statistically expected into the DSB site. The process can be further enhanced if the T-DNA is flanked by sequences homologous that are homologous to the genomic target site (“gene targeting”). More recently, we were able to set up the so-called “in planta” gene targeting (GT) technique. Here, the targeting vector is first integrated into the genome and is then excised at the same time as the DSB is induced in the target locus. Thus, GT occurs during the life cycle of a plant and targeted events can be harvested as seeds. No mutagenic and labor intensive tissue culture is necessary with this kind of technique. Using the CRISPR/Cas system (see below), we were able to demonstrate that the technique is applicable for targeting natural genes in Arabidopsis (see Figure 1).
Figure 1: Principle of the in planta gene targeting system. Upon expression of the stably transformed CRISPR/Cas nuclease, three DSBs are introduced: one is formed in the desired target locus, activating it for homologous recombination (HR). The other two the so-called GT vector, which is located on the same T-DNA as the nuclease. The transgene on the GT vector then integrates into the activated target locus by HR, using the flanking homologies. The result is a site-specific and heritable integration of the transgene.
Since the discovery of CRISPR/Cas system as a sequence-specific endonuclease that can be easily programmed to cut any specific genome sequence at will using a simple guide RNA, genome engineering is in full bloom in various eukaryotes. We were able to show that the system can be used with great ease to induce heritable mutations in natural genes in Arabidopsis. We also could show that by the use of a paired nickase instead of a nuclease, mutagenesis can be performed avoiding off-site effects in the genome of Arabidopsis.
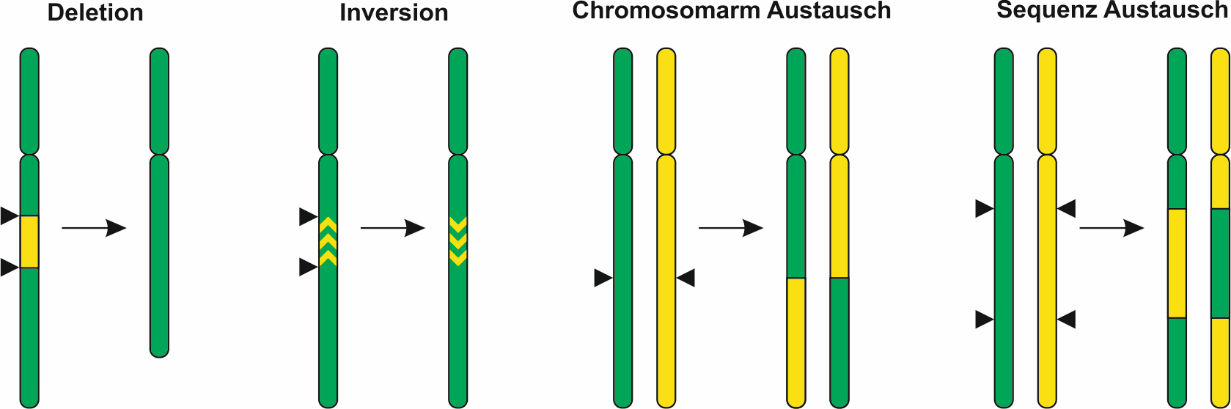
Currently, we are in the process of establishing a number of further applications of the CRISPR/Cas system for genome engineering such as inversions or translocations (see Figure 2). In the long run, CRISPR/Cas will pave the way to the synthetic plant genome.
Figure 2: Applications for genomic engineering in plants. Induction of two DSBs within a chromosome may lead to deletions or inversions. Induction of two DSBs on different chromosomes may lead to the exchange of chromosome arms. Induction of four DSBs may be used to exchange sequences between chromosomes.
HUMAN GENETIC DISORDERS AND BREAST CANCER GENES AS KEYS TO GENOME STABILITY AND INHERITANCE IN PLANTS
The second focus of our research concentrates on the understanding of the complex network of proteins involved in DNA repair and DNA recombination in plants. Whereas the identification of factors with functions in DNA recombination similar to other organisms was a center of interest before, we more recently focused our attention on plant-specific peculiarities and on defining the individual roles of the different factors in multiple DNA repair pathways of plants.
Factors related to human genetic disease syndromes
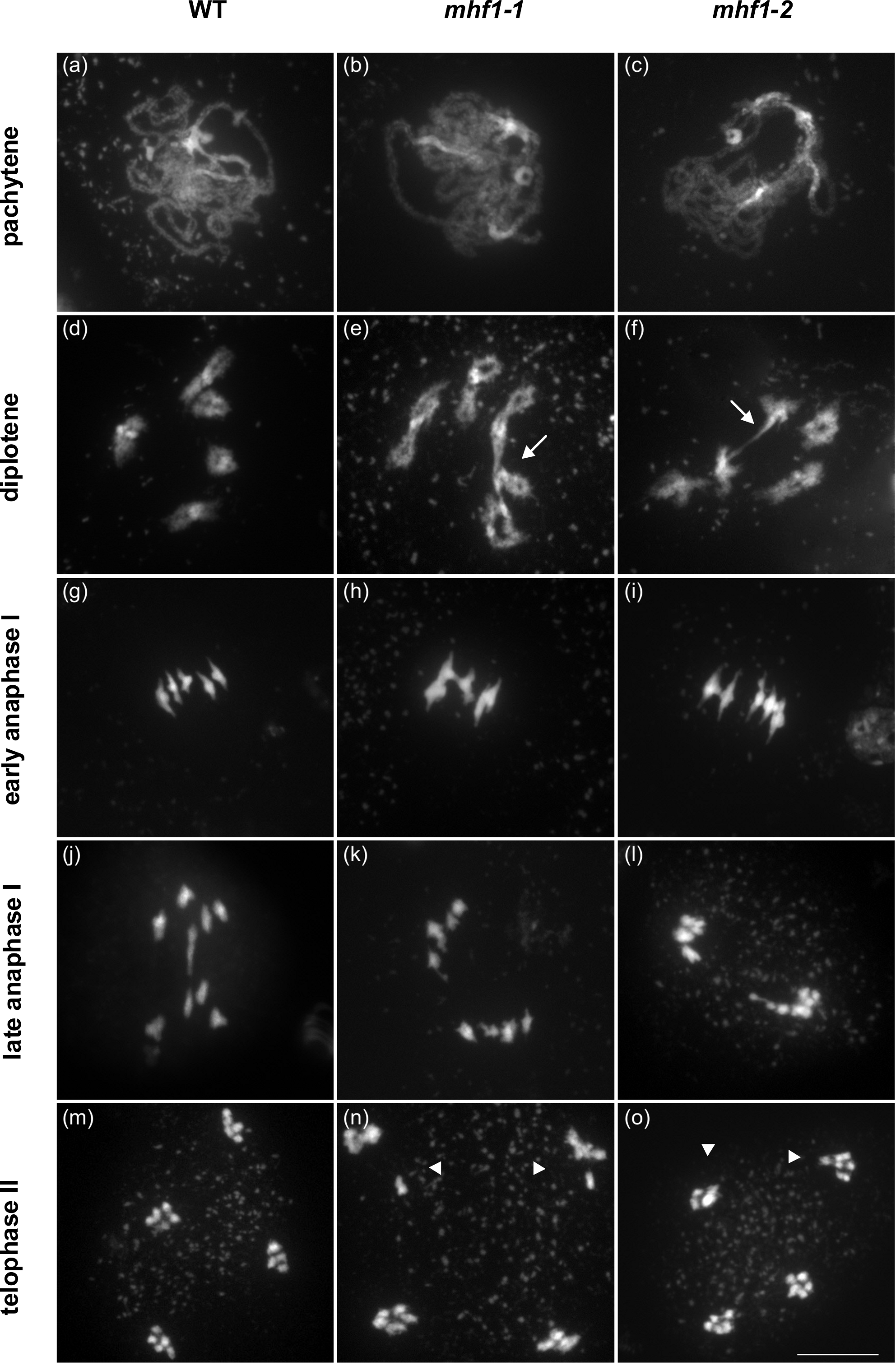
In humans the genetic disorders Bloom syndrome (BS) and Werner syndrome (WS) are caused by mutations in genes that are coding for RecQ family DNA helicases. Patients with Werner syndrome show accelerated senescence and a high prevalence of cancer. In Bloom syndrome patients, a higher incidence of cancer is also found, in addition to elevated rates of sister chromatid exchanges at the cellular level. We were able to identify a surprisingly large family of seven RecQ homologues in Arabidopsis. By analyzing insertion mutants and CRISPR/Cas induced mutations, we characterize their biological function in vivo. Our results indicate that the AtRECQ4A mutants show sensitivity against DNA damaging agents and exhibit enhanced homologous recombination (HR) rates. Together with genetic interactions with other factors, these facts strongly suggest that RECQ4A is functionally equivalent to the mammalian BLM protein. Fanconi anemia (FA) is an inherited disease leading to different symptoms like bone marrow failure, congenital abnormalities and cancer. The affected cells exhibit an increased sensitivity against interstrand crosslink (CL)-inducing agents. Interestingly, most FA genes are only weakly conserved between eukaryotes. We analyze the biological role of several homologues in Arabidopsis. Few FA homologues have such a prominent role in CL repair in plants as they have in mammals. We could show that the helicase AtFANCM is involved in interstrand CL repair and also in the suppression of HR somatic cells. Surprisingly, and in contrast to the animal and yeast homologues, AtFANCM and AtMHF1 seems also to be required for proper meiotic recombination (see Figure 3).
Figure 3: Chromatin spreads of Arabidopsis thaliana cells undergoing meiosis. Exemplary meiocytes of wild-type (WT; a, d, g, j, m), mhf1-1 (b, e, h, k, n) and mhf1-2 (c, f, i, l, o) cells. In the pachytene stage (a–c), neither mhf1 mutant exhibited defects compared with WT meiocytes. In the diplotene stage (d–f), connections between the five bivalents in mhf1-1 and mhf1-2 were observed (arrows). In early anaphase I (g–i), those bridges were also apparent, resulting in unequal chromosome distributions in both mhf1 mutants at the end of the second meiotic division in telophase II (m–o, arrowheads). Scale bar = 10 µm.
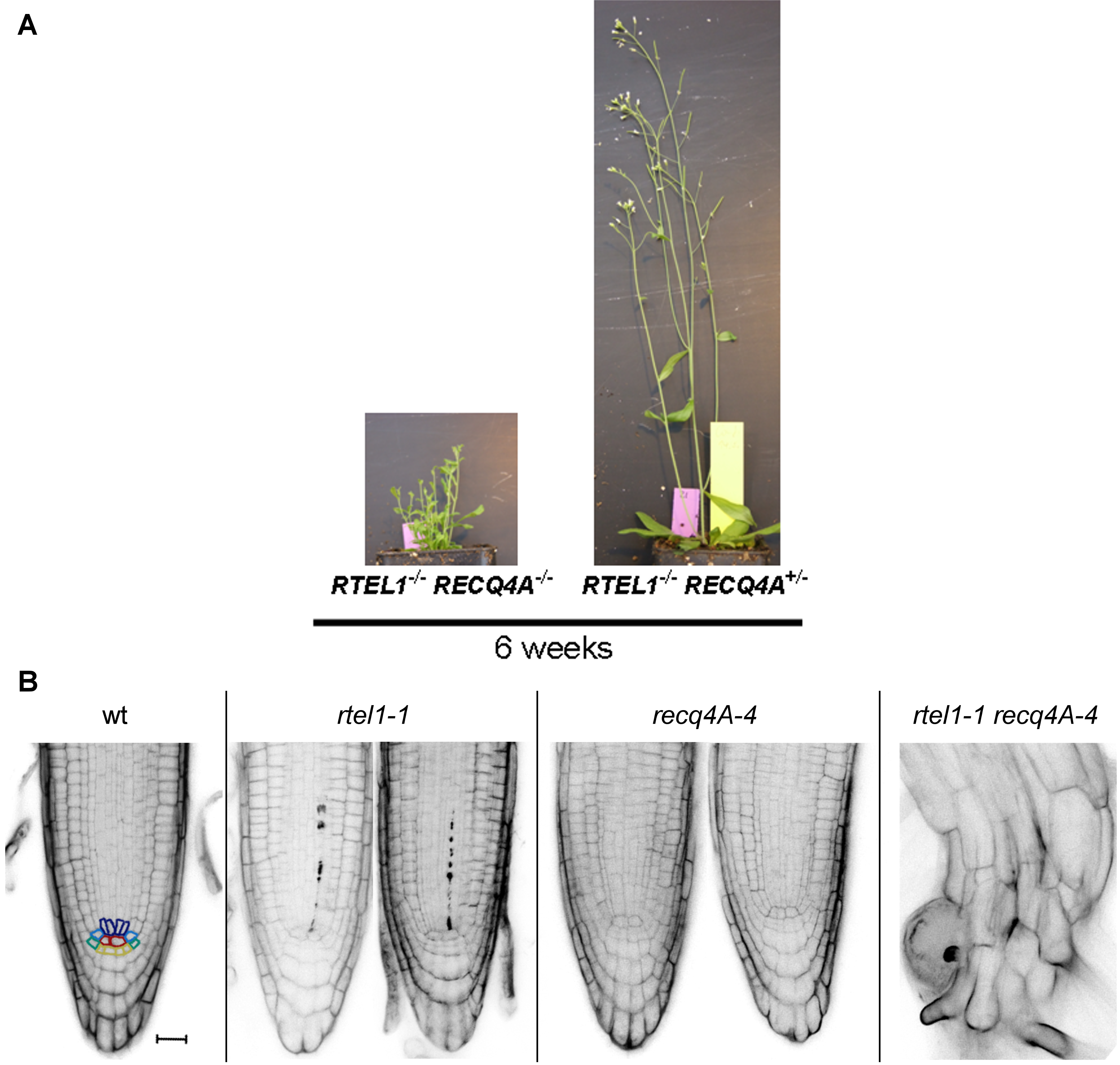
In humans, mutations in the DNA helicase “Regulator of Telomere Elongation Helicase1” (RTEL1) leads to Hoyeraal-Hreidarsson syndrome, a severe, multisystem disorder. We could demonstrate that the RTEL1 homologue of Arabidopsis is not only involved in the repair of CLs and the stabilization of telomeres, but also in the suppression of HR. Interestingly, all three helicases that are involved in the suppression of HR, AtRTEL1, AtRECQ4A and AtFANCM, respectively, operate in independent pathways, demonstrating how important the regulation of HR is for genome stability in plants. That AtRTEL1 and AtRECQ4A are involved in independent pathways of replicative DNA repair is also exemplified by the strong growth phenotype of the double mutant (see Figure 4).
Figure 4: Severe Growth Defect Due to the Concurrent Loss of RTEL1 and RECQ4A. (A) Loss of both helicases led to smaller rosette leaves, shorter shoots, and the absence of true flowers or siliques in the rtel1-1 recq4A-4 double mutant. (B) Furthermore PI-stained root tips of the double mutant had severely short roots with a complete loss of root structure. Scale bar = 20 mm.
Factors homologous to genes involved in human breast cancer
Surprisingly, plants carry homologues of genes that are involved in the suppression of breast cancer in humans. Mutations of AtBRCA1and the two AtBRCA2 genes lead to a deficiency in HR. Currently we are in the process of defining genetic interaction networks of these proteins to learn more about the reaction of plants to genotoxic stress.
Biochemical characterization of DNA processing enzymes
Many factors that are required for DNA repair and DNA recombination are directly involved in the processing of DNA molecules. Beside analyzing their biological role in vivo, we also characterize their biochemical properties in vitro. In the center of our interest are the RecQ helicases and the translocase AtRAD5A, nucleases that process recombination intermediates like AtMUS81, AtSEND1 and AtGEN1 as well as DNA binding proteins like Replication Protein A homologues.
Inheritance – from understanding to directing meiotic recombination
Our research revealed that several topoisomerase-related factors have a decisive role in meiotic recombination. We were able to identify three homologues of Spo11, the protein responsible for the induction of DSBs during meiosis, which is encoded by a single gene in other eukaryotes. Spo11 descends from the subunit A of the archaebacterial topoisomerase VI. We were able to demonstrate that plants carry - in contrast to other eukaryotes - also a B subunit of this enzyme. This subunit together with one of the three Spo11 homologues constitutes a topoisomerase that is mainly required for DNA endoreduplication in somatic plant cells. Furthermore, we were able to show that the two other Spo11 homologues are both actively involved in meiotic DSB induction.
We were also able to discover a previously unanticipated way of decatenation of meiotic recombination intermediates. We were able to show that the topoisomerase TOP3α as well as the structural protein RMI1 are essential for the proper progression of meiotic recombination in Arabidopsis. Interestingly, recent results of other groups indicate that this step is also part of the meiotic recombination reactions in other eukaryotes, indicating the power of plants as an experimental system to study meiotic recombination.
As plant breeding is based on meiotic recombination and the distribution of crossovers is decisive for the combination of beneficial traits and the breaking of linkages in case of adverse traits, our efforts at the moment concentrate on the question whether we can control the respective steps by means of genetic engineering.