CRISPR/Cas Plasmids & Protocols for Genome Engineering in Plants
We demonstrated that:
On the gene level
- Heritable targeted mutagenesis events can be induced using the SpyCas9, SauCas9 and SthCas9 nucleases
- Heritable homologous recombination based gene targeting events can be achieved by combining the SpyCas9 with our previously developed in planta gene targeting technology
- Tandem duplications can be induced by using paired SpyCas9 nickases
- Catalytic inactive versions of the SauCas9 nuclease and and SpyCas9 fused to fluorescent proteins can be used for live cell imaging of telomeric regions in plants
- LbCas12a can be engineered for highly efficient temperature-tolerant plant gene editing (ttLbCas12a)
- The editing efficiency of ttLbCas12 can be further enhanced by the inclusion of introns (ttLbCas12a-i)
- In planta gene targeting can be further improved by using egg-cell specific expression of SauCas9, ttLbCas12a and ttLbCas12a-i
- Efficiency of SpyCas9-mediated mutagenesis and size of induced deletions can be increased by recruiting the exonuclease TREX1
- In planta gene targeting frequencies are increased in cNHEJ-deficient ku70 mutants
- The nuclease ErCas12a can be engineered for efficient temperature-tolerant plant gene editing (imErCas12a)
On the chromosome level
- Heritable chromosomal translocations can be induced efficiently by using egg-cell specific expression of the SauCas9 nuclease
- These translocation can be used to break genetic linkages for breeding
- Heritable chromosomal inversion can be induced efficiently by using egg-cell specific expression of the SauCas9 nuclease
- These inversion can be used to lock or unlock meiotic recombination between homologous chromosomes
- SauCas9 can be used for organ specific cell elimination in plants
- Coupling the CRISPR-Kill system with a chemically inducible expression system enables temporal control of CRISPR-mediated cell death induction
- The number of chromosomes can be changed through chromosome fusion
Publications:
- Rönspies, Michelle; Khosravi, Solmaz; Helia, Ondřej; Valisi, Alessandro, Fajkus, Jiří; Fojtová, Miloslava; Houben, Andreas; Puchta, Holger (2025): CRISPR-Cas-mediated heritable chromosome fusions in Arabidopsis. In: Science 390 (6775), S. 843-848. DOI: 10.1126/science.adz8505.
- Pietralla, Janine; Capdeville, Niklas; Schindele, Patrick; Puchta, Holger (2024): Optimizing ErCas12a for efficient gene editing in Arabidopsis thaliana. In: Plant Biotechnology Journal, Artikel pbi.14194. DOI: 10.1111/pbi.14194.
- Merker, Laura; Feller, Laura; Dorn, Annika; Puchta, Holger (2024): Deficiency of both classical and alternative end-joining pathways leads to a synergistic defect in double-strand break repair but not to an increase in homology-dependent gene targeting in Arabidopsis. In: The Plant journal. DOI: 10.1111/tpj.16604.
- Capdeville, Niklas; Schindele, Patrick; Puchta, Holger (2023): Increasing deletion sizes and the efficiency of CRISPR/Cas9-mediated mutagenesis by SunTag-mediated TREX1 recruitment. In: The Plant journal. DOI: 10.1111/tpj.16586.
- Gehrke, Fabienne; Ruiz‐Duarte, Paola; Schindele, Angelina; Wolf, Sebastian; Puchta, Holger (2023): An inducible CRISPR‐Kill system for temporally controlled cell type‐specific cell ablation in Arabidopsis thaliana. In: New Phytologist, Artikel nph.19102. DOI: 10.1111/nph.19102.
- Schindele, Patrick; Merker, Laura; Schreiber, Tom; Prange, Anja; Tissier, Alain; Puchta, Holger (2022): Enhancing gene editing and gene targeting efficiencies in Arabidopsis thaliana by using an intron‐containing version of tt Lb Cas12a. In Plant Biotechnology Journal, Article pbi.13964. DOI: 10.1111/pbi.13964.
- Rönspies, Michelle; Schmidt, Carla; Schindele, Patrick; Lieberman-Lazarovich, Michal; Houben, Andreas; Puchta, Holger (2022): Massive crossover suppression by CRISPR–Cas-mediated plant chromosome engineering. In Nature plants. DOI: 10.1038/s41477-022-01238-3.
- Schindele, Angelina; Gehrke, Fabienne; Schmidt, Carla; Röhrig, Sarah; Dorn, Annika; Puchta, Holger (2022): Using CRISPR-Kill for organ specific cell elimination by cleavage of tandem repeats. In Nat. Commun. 13 (1502). DOI: 10.1038/s41467-022-29130-w
- Wolter, Felix; Schindele, Patrick; Beying, Natalja; Scheben, Armin; Puchta, Holger (2021): Different DNA repair pathways are involved in single-strand break-induced genomic changes in plants. In The Plant cell. DOI: 10.1093/plcell/koab204
- Huang, Teng-Kuei; Armstrong, Brittney; Schindele, Patrick; Puchta, Holger (2021): Efficient gene targting in Nicotiana tabacum using CRISPR/SaCas9 and temperature tolerant LbCas12a. In: Plant Biotechnology J. DOI: 10.1111/pbi.13546.
- Schmidt, Carla; Fransz, Paul; Rönspies, Michelle; Dreissig, Steven; Fuchs, Jörg; Heckmann, Stefan; Houben, Andreas & Puchta, Holger (2020): Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. In: Nat. Commun 11, 4418. DOI: 10.1038/s41467-020-18277-z
- Beying, Natalja; Schmidt, Carla; Pacher, Michael; Houben, Andreas; Puchta, Holger (2020): CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. In: Nat. Plants 19, S. 778. DOI: 10.1038/s41477-020-0663-x
- Merker, Laura; Schindele, Patrick; Huang, Teng-Kuei; Wolter, Felix; Puchta, Holger (2020): Enhancing in planta gene targeting efficiencies in Arabidopsis using temperature-tolerant CRISPR/LbCas12a. In: Plant biotechnology journal. DOI: 10.1111/pbi.13426
- Schindele, Patrick; Puchta, Holger (2019): Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. In: Plant biotechnology journal. DOI: 10.1111/pbi.13275
- Wolter, Felix; Puchta, Holger (2019): In planta gene targeting can be enhanced by the use of CRISPR /Cas12a. In: Plant J. DOI: 10.1111/tpj.14488
- Schmidt, Carla; Pacher, Michael; Puchta, Holger (2019): Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. In: The Plant journal : for cell and molecular biology. DOI: 10.1111/tpj.14322
- Wolter, F. , Klemm, J. and Puchta, H. (2018): Efficient in planta gene targeting in Arabidopsis using egg‐cell specific expression of the Cas9 nuclease of S. aureus. Plant J. Accepted Author Manuscript. . DOI:10.1111/tpj.13893
- Vu, G. T., Cao, H. X., Fauser, F., Reiss, B., Puchta, H. & Schubert, I. (2017): Endogenous sequence patterns predispose the repair modes of CRISPR/Cas9‐induced DNA double strand breaks in Arabidopsis thaliana. The Plant Journal
- Dreissig, S., Schiml, S., Schindele, P., Weiss, O., Rutten, T., Schubert, V., Gladilin, E., Mette, MF., Puchta, H. & Houben, A. (2017): Live cell CRISPR‐imaging in plants reveals dynamic telomere movements. The Plant Journal
- Schiml, S., Fauser, F., & Puchta, H. (2016): Repair of adjacent single-strand breaks is often accompanied by the formation of tandem sequence duplications in plant genomes. Proceedings of the National Academy of Sciences, 201603823
- Steinert J., Schiml S., Fauser F. and Puchta H. (2015): Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. DOI: 10.1111/tpj.13078
- Schiml S., Fauser F. and Puchta H. (2014): The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. DOI: 10.1111/tpj.12704
- Fauser F., Schiml S. and Puchta H. (2014): Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana.Plant J 79, 348-359
Reviews:
- Yaşar, Seda; Gehrke, Fabienne; Capdeville, Niklas; Puchta, Holger (2026): Recent progress in plant genome engineering: from large insertions to chromosome number changes. In: Current Opinion in Biotechnology 97, S. 103426. DOI:10.1016/j.copbio.2025.103426.
- Gilbertson, Larry; Puchta, Holger; Slotkin, R. Keith (2025): The future of genome editing in plants. In: Nat. Plants DOI: 10.1038/s41477-025-01956-4.
- Puchta, Holger; Houben, Andreas (2024): Plant chromosome engineering – past, present and future. In: New Phytologist. DOI: 10.1111/nph.19414.
- Capdeville, Niklas; Schindele, Patrick; Puchta, Holger (2023): Getting better all the time — recent progress in the development of CRISPR/Cas-based tools for plant genome engineering. In: Current Opinion in Biotechnology 79, S. 102854. DOI: 10.1016/j.copbio.2022.102854.
- Rönspies, Michelle and Puchta, Holger (2022): Redirecting meiotic recombination by CRISPR-Cas-mediated chromosome engineering. In Nature plants. DOI: 10.1038/s41477-022-01239-2.
- Gehrke, Fabienne; Schindele, Angelina; Puchta, Holger (2022): Nonhomologous end joining as key to CRISPR/Cas-mediated plant chromosome engineering. In Plant physiology 188 (4), pp. 1769–1779. DOI: 10.1093/plphys/kiab572.
- Puchta, Holger; Jiang, Jiming; Wang, Kan; Zhao, Yunde (2022): Updates on gene editing and its applications. In Plant physiology 188 (4), pp. 1725–1730. DOI: 10.1093/plphys/kiac032.
- Rönspies, Michelle; Dorn, Annika; Schindele, Patrick; Puchta, Holger (2021): CRISPR-Cas-mediated chromosome engineering for crop improvement and synthetic biology. In Nature plants 7 (5), S.566–573. DOI: 10.1038/s41477-021-00910-4.
- Huang, Teng-Kuei and Puchta, holger (2021): Novel CRISPR/Cas applications in plants: from prime editing to chromosome engineering. In: Transgenic Res. DOI: 10.1007/s11248-021-00238-x.
- Capdeville, Niklas; Merker, Laura; Schindele, Patrick; Puchta, Holger (2021): Sophisticated CRISPR/Cas tools for fine-tuning plant performance. In: Journal of Plant Physiology. 257, S. 153332. DOI: 10.1016/j.jplph.2020.153332.
Protocols:
- Gehrke, Fabienne; Wolf, Sebastian; Puchta, Holger (2024): Protocol to efficiently induce CRISPR-Kill-mediated cell ablation in Arabidopsis thaliana. In: STAR Protocols 5 (2), 103072. DOI: 10.1016/j.xpro.2024.103072.
- Rönspies, Michelle; Schindele, Patrick; Wetzel, Rebecca; Puchta, Holger (2022): CRISPR–Cas9-mediated chromosome engineering in Arabidopsis thaliana. In Nat Protoc. DOI: 10.1038/s41596-022-00686-7.
- Merker, L.; Schindele, P. & Puchta, H. (2020): Using CRISPR/ttLbCas12a for in planta gene targeting in A. thaliana. In: Current Protocols in Plant Biology, 5, e20117. DOI: 10.1002/cppb.20117
- Schindele, P.; Wolter, F. & Puchta, H. (2020): CRISPR Guide RNA Design Guidelines for Efficient Genome Editing. In: Methods in molecular biology (Clifton, N.J.) 2166, S. 331–342. DOI: 10.1007/978-1-0716-0712-1_19
- Khosravi, S.; Dreissig, S.; Schindele, P.; Wolter, F.; Puchta, H. & Houben, A. (2020): Live-Cell Imaging in Plant Cells with a Telomere-Specific Guide RNA. In: Methods in molecular biology (Clifton, N.J.) 2166, S. 343-356. DOI: 10.1007/978-1-0716-0712-1_20
- Wolter, Felix; Huang, Teng-Kuei; Puchta, Holger (2020): Efficient Homologous Recombination-Mediated in Planta Gene Targeting by Egg-Cell-Specific Expression of Staphylococcus aureus Cas9 from Arabidopsis. In: M. Tofazzal Islam, Pankaj K. Bhowmik und Kutubuddin A. Molla (Hg.): CRISPR-Cas Methods. New York, NY: Springer US, S. 25–34. DOI 10.1007/978-1-0716-0616-2_2
- Steinert, J., Schmidt, C. & Puchta, H. (2017): Use of Cas9 Orthologs from Streptococcus thermophilus und Staphylococcus aureus for Non-Homologous End-Joining Mediated Site-Specific Mutagenesis in Arabidopsis thaliana Plant Gerline Development: Methods and Protocols, Methods in Molecular Biology, vol. 1660, DOI 10.1007/978-1-4939-7286-9_27
- Schiml, S., Fauser, F. & Puchta, H. (2017): CRISPR/Cas-Mediated In Planta Gene Targeting. Plant Genomics: Methods and Protocols, 3-11.
- Schiml S., Fauser F. & Puchta H. (2016):CRISPR/Cas-Mediated Site-Specific Mutagenesis in Arabidopsis thaliana Using Cas9 Nucleases and Paired Nickases Chromosome and Genomic Engineering in Plants In: Methods and Protocols, Methods in Molecular Biology, vol. 1469, DOI 10.1007/978-1-4939-4931-1_8
Cloning Protocol
Here, we describe our Gateway-compatible cloning protocol for single sgRNAs. To obtain any of the available plasmids or if there are questions about the procedure, please contact us directly.
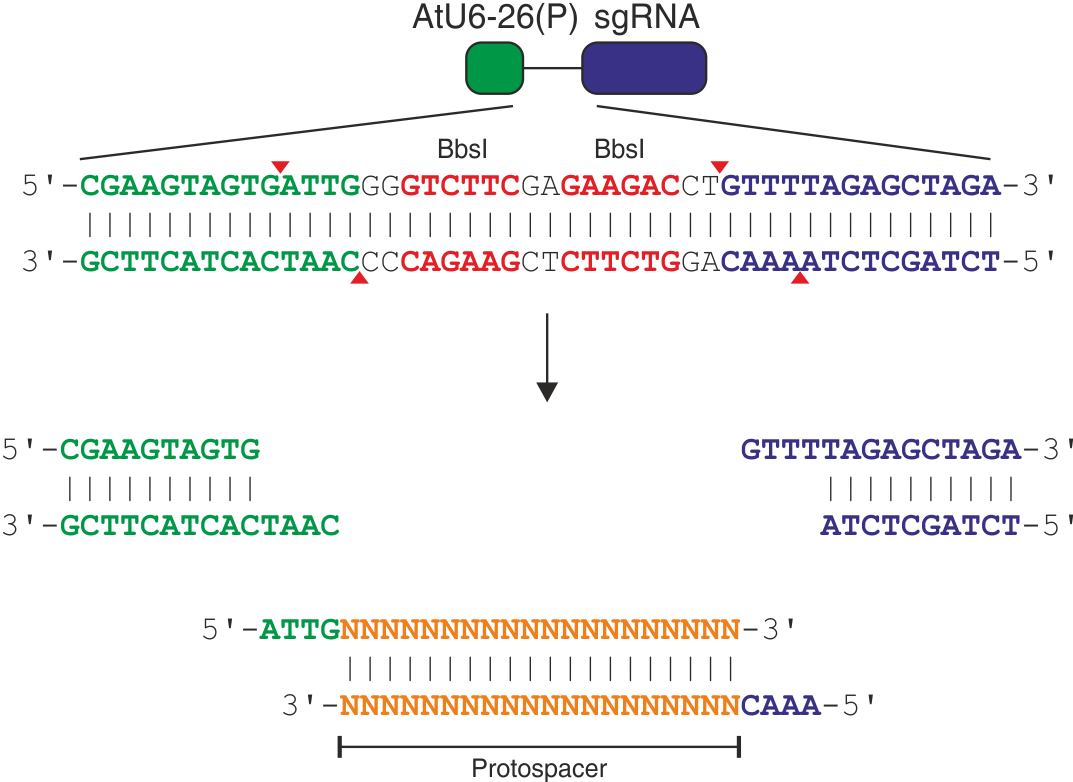
- Preparations
- Pick your 20-nt protospacer sequence. For detailed information refer to the respective orthologue you are using.
- Order desalted oligos:
- FW: 5'-ATTG + protospacer
- REV: 5'-AAAC + rev-com protospacer
- Oligo annealing
- 2 µl of each oligo (50 µM) + 46 µl ddH20
- Incubate for 5 min, 95°C (thermocycler, no cooling at the end!)
- Cooling at RT for 20 min
- Digest entry vector
- 10 µl pEn-Chimera
2 µl 10x NEB buffer 2.1
1 µl NEB BbsI (stored at -80°C!)
7 µl ddH2O - Incubate for >1 h, 37°C
- Purify
- Adjust to 5 ng/µl
- 10 µl pEn-Chimera
- Ligation
- 2 µl of digested pEn-Chimera
3 µl of annealed oligos
1 µl of T4 Ligase
5 µl of NEB 2x Quick Ligase Buffer (or any other T4 buffer) - Incubate for 1h, RT
- Transform 5 µl in NEB5alpha, plate 100 µl on LB/Amp
- 2 µl of digested pEn-Chimera
- Colony-PCR
- Test 5 colonies (efficiency >70%)
- Use fw-oligo + SS129 as primers
- Anneal at 56°C, 30 s elongation, 30 cycles
- Expected band at 370 bp
- Miniprep
- Sequence using primer SS42
- Adjust to 100 ng/µl
- Gateway Reaction
- 2 µl of your entry vector
3 µl of respective CAS9 vector (adjust to 50 ng/µl)
4 µl TE buffer, pH 8
1 µl LR clonase II - Vortex + centrifuge
- Incubate for >2 h, RT
- Proteinase K treatment: add 1 µl; incubate for 10 min, 37°C (crucial step!)
- Transform completely in NEB5alpha, plate 100 µl on LB/Spec
- 2 µl of your entry vector
- Optional: Colony-PCR
- All colonies should be OK, test 3-6
- Use primers SS42 / SS43
- Anneal at 60°C, 1 min elongation, 35 cycles
- Expected band at 1070 bp
- For vectors with Kanamycin plant selection:
- Use primers SS42 / SS 102
- Anneal at 60 °C, 1 min elongation, 35 cycles
- Expected band at 917 bp
- Miniprep
- Control by digestion using AflII and NheI
- Expected bands (nuclease): 5.9 kb, 5.0 kb and 3.8 kb
- Optional: Sequence using SS42 and SS61
- Control by digestion using AflII and NheI
- Primer Sequences
- SS42: TCCCAGGATTAGAATGATTAGG
- SS43: CGACTAAGGGTTTCTTATATGC
- SS61: GAGCTCCAGGCCTCCCAGCTTTCG
- SS129 (M13): CACAGGAAACAGCTATGAC
- SS102 CACCATGTTATCACATCAATCC
Plasmids
We are happy to share all of our CRISPR/Cas reagents with the plant community. Please feel free to contact us directly if you are interested in any of our reagents. If you are interessted in our plasmids concerning the different Cas9 othologs and Cas12a you can find them in our special sections here: LbCas12a, S.pyogenes, S.aureus, S.thermophilus

